
NETwork! analysis in cancer – managing genomics research data and building a repository workflow
Professor Cristin Print, Dr Nicholas Knowlton, Faculty of Medical and Health Sciences
NETwork!
A New Zealand wide alliance of cancer clinicians and scientists who are working together in a new national framework to manage and study neuroendocrine cancers. The research team consist of multiple disciplinary domains including medical oncology, neuroendocrine tumours (NETs), genomics and bioinformatics, molecular biology, next generation sequencing and pharmacology. It is based out of the clinical trials coordinating centre, and Auckland and Christchurch Tissue Banks, and incorporates surgeons from numerous specialty areas, District Health Boards across New Zealand and several professional Community engagement teams.
The NETwork project, based in the Faculty of Medical and Health Sciences, aims to record information on all patients diagnosed with NETs in New Zealand. Another aim is to identify markers for the identification and follow-up of NETs using non-invasive testing methods such as blood tests. All analyses are to help service providers to make better treatment decision, to guide patient care, and to speed the time to get a diagnosis for patients.
Initially, the NETwork team followed a small-scale data management plan – appropriate for the small size of project when it started. However, the group has rapidly grown and needs to mature the data management processes from managed file systems to a structured repository with well documented, automated workflows.
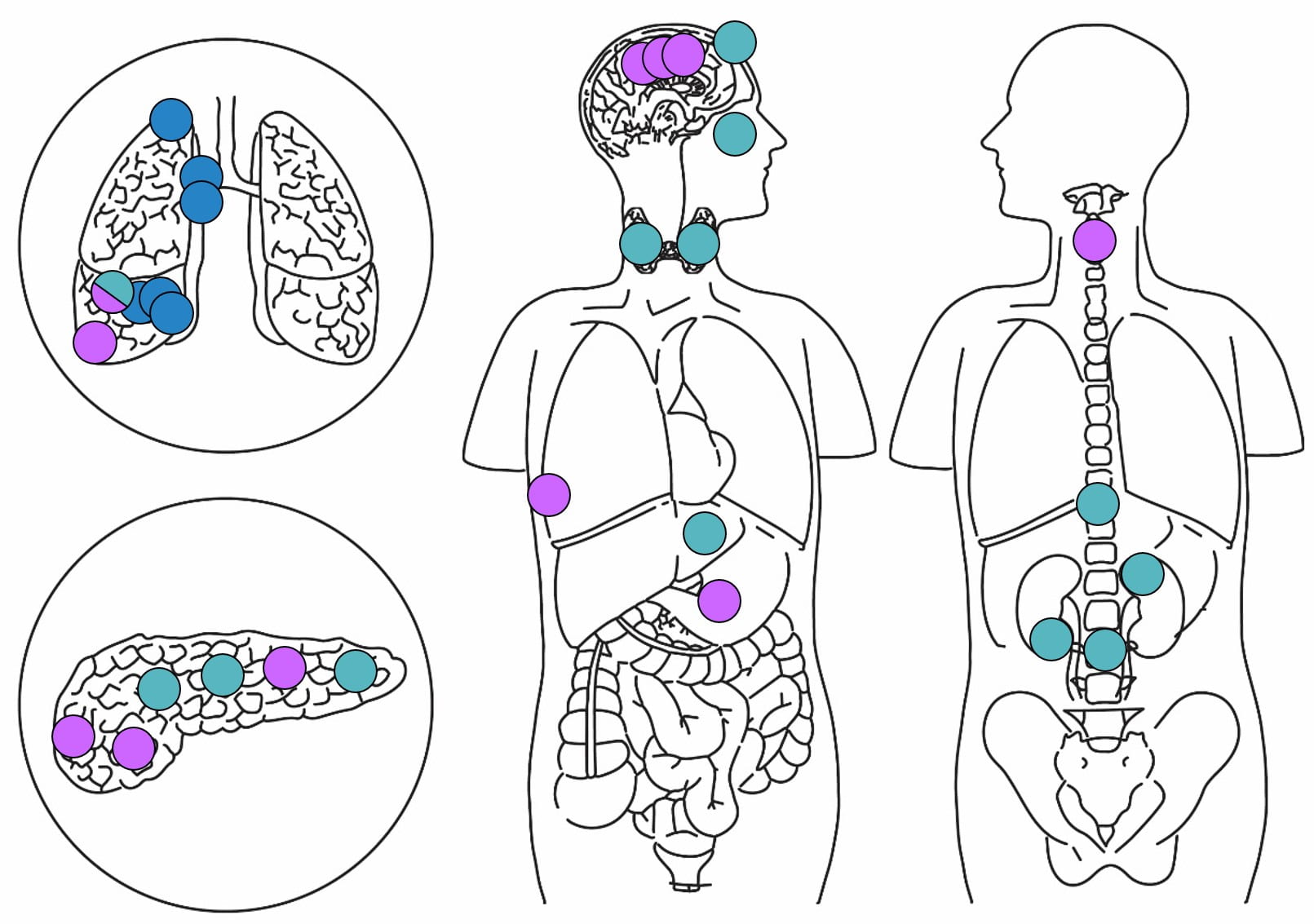
Figure 1. The two genomically distinct distant metastatic lineages (coloured as above) are spatially dispersed around the patient’s body. (T.Robb, C. Blenkiron, P. Tsai K. Parker, B. Woodhouse, B. Lawrence, C. Prin etc., “Investigating Tumour Evolution in a Single Disseminated Cancer”, Poster, NZSO and Queenstown Research Week 2018.)
MyTardis data repository
The Centre for eResearch (CeR) is currently building a genomics data repository for the NETwork research program. Currently we are facilitating semi-automated processing of data into an instance of MyTardis, a data repository system developed at Monash University. Doing so allows for secure and regularly backed-up storage of genomics data alongside clinical metadata into a managed system. Next steps are to work towards an automated workflow of data to and from the NZ eScience Infrastructure (NeSI) HPC platform for bioinformatics analysis, return of the results generated into the MyTardis repository, and further analysis, including data visualisation, on the Nectar research cloud using the Genomics Virtual Lab (GVL).
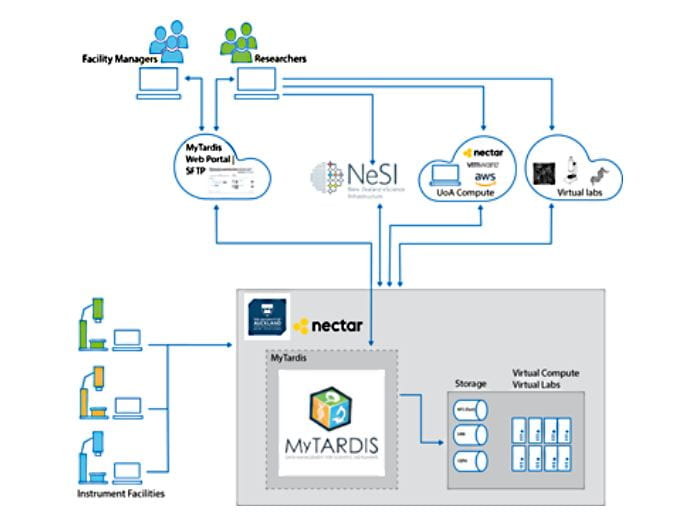
MyTardis workflow
The GVL
A pre-configured (Figure 2) platform running on Nectar cloud infrastructure, which provides tools for large-scale analysis and visualisation of next generation sequencing data. The genomics group will work with CeR to expand the NETwork pilot to all medical genomics researchers within UoA, providing a substantial contribution to managing the risks inherent in data generated by human genomics research, which is currently handled in an ad-hoc manner, while facilitating access to this data to authorised researchers.
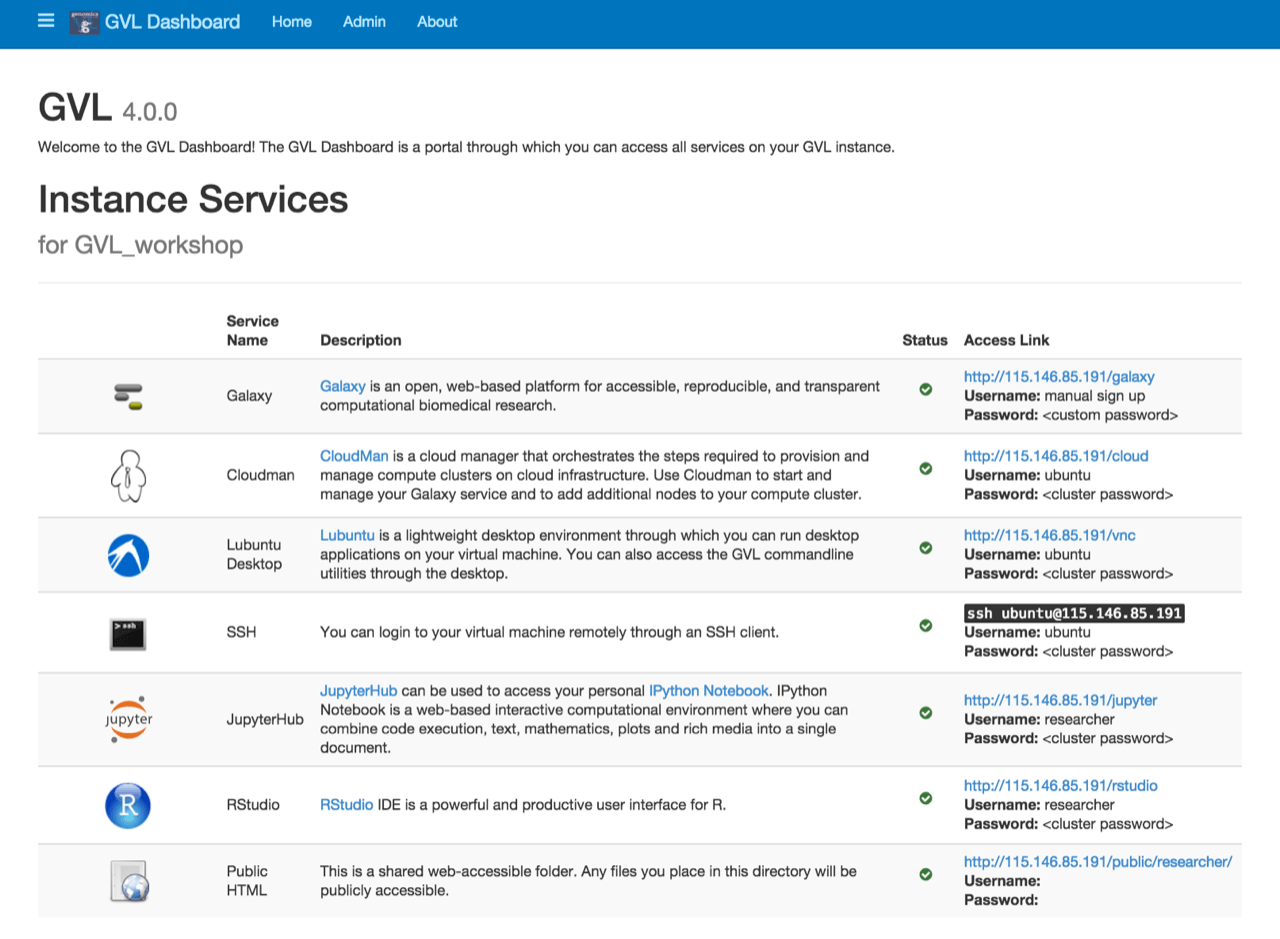
Figure 2. GVL dashboard which lists various instane servies it can run from Nectar.
It is imperative for UoA to develop a fit-for-purpose research infrastructure, and promote best practice in data management amongst its research community. The University acknowledges its obligations, and the necessity to achieve compliance with this policy framework as well as the funder’s data policy to ensure long-term sustainability.
UoA subscribes to a model of data management that strives to meet FAIR data standards. These can be summed up as: ‘making data as accessible as possible and as closed as necessary’. The acronym FAIR refers to data being: Findable, Accessible, Interoperable and Reusable. Given the nature of the data being stored within the repository, the CARE principles of data management also apply, particularly in the context of indigenous data.CARE is an extension to FAIR and sets principles of Collective benefit, Authority to control, Responsibility and Ethics to the data management plan, in addition to the FAIR principles general to all data.
The larger MyTardis genomics in medicine (MyGIM) data repository (part of the second phase of the Genomics in Medicine Strategic Research Initiative Fund) will build on FAIR data principles and provide the University with the capability to facilitate new data science research using medical genomics data generated at the University. In the coming year, CeR will provide seminars and training, and in some cases one-on-one mentorship, in data science related to genomics within AAHA. he overall intention is to raise data science literacy in genomics clinicians and researchers to meet the challenge of future computational genomics literacy. Relatively little of the medical genomics data currently generated in the University is utilised in data science research, which is a lost opportunity for our researchers, and ultimately for New Zealand patients, to better understand health and disease. By leveraging FAIR data principles whilst lowering the barriers to accessing computational resources and providing appropriate training, CeR is helping to address this lost opportunity.